Syntax Builder¶
What is the OpenCloning Syntax Builder?¶
The OpenCloning Syntax Builder is a tool that allows you to create and edit syntaxes for your own cloning kits.
How to use the OpenCloning Syntax Builder¶
Go to https://syntax.opencloning.org/, which will offer a list of options to start from. If your syntax is based on an existing one, you can click on Loading an example. If you want to start from scratch, we recommend starting by Defining parts from overhangs, but we show the examples below for Defining parts directly.
Let's take as an example the syntax of SubtiToolKit, which is a kit for B. subtilis and Gram-positive bacteria. The assemblies of Transcriptional Units (TUs) are displayed below:
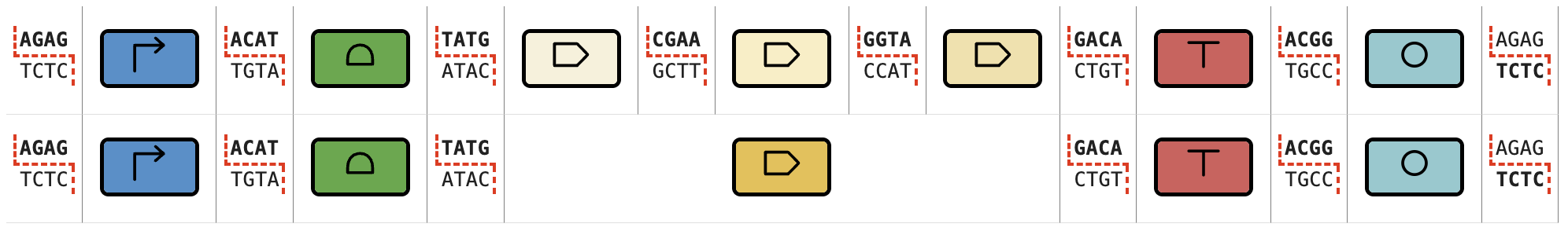
We can see that there are two "paths", one where we include a single CDS, and another where we include up to three CDS.
Defining parts from overhangs¶
In this case, you simply provide the overhangs in order as they appear in the syntax. For our example, we would write:
AGAG
ACAT
TATG
GACA
ACGG
AGAG
TATG
CGAA
GGTA
GACA
- The first block includes one overhang per line for the "main path", and ends at the starting point to indicate that the assembly is circular.
- Then a blank line to define a "branching path", starting from the branching point
TATGand ending where the branch merges again,GACAin this case. - More branching paths can be added if needed by adding a blank line and repeating the process.
In this particular example, we could have just defined a single block with the small steps, and added the "one CDS" TATG-GACA as a part later on (see next section).
AGAG
ACAT
TATG
CGAA
GGTA
GACA
ACGG
AGAG
Once you are done, click on Next, and it will take you define parts (see next section).
Defining parts directly¶
You can start from a blank slate, or generate a few parts from overhangs as below. In either case, you can edit / add / remove parts in this interface:
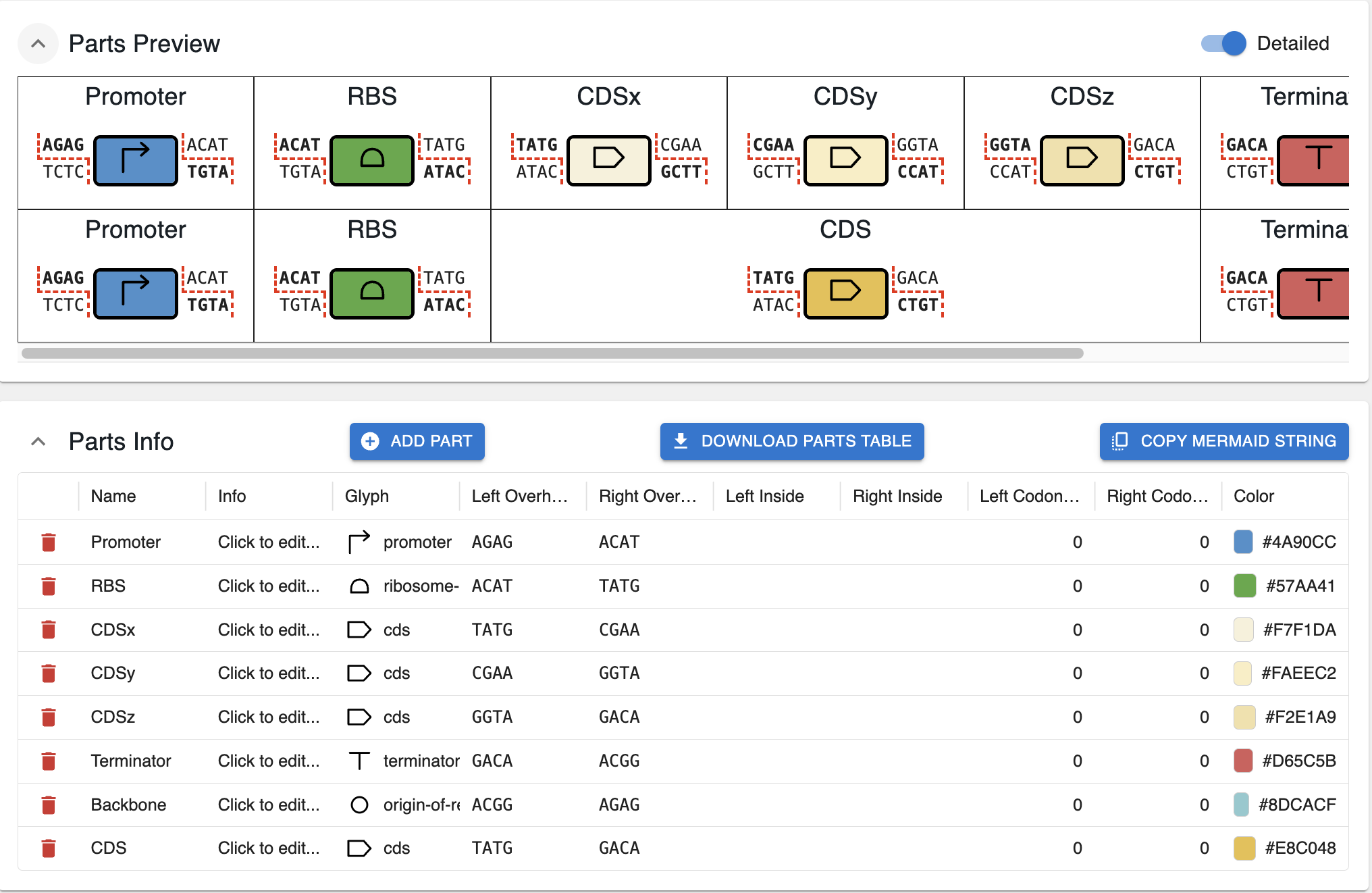
For each part, you can define:
- Name: the human‑readable name of the part (e.g.
Promoter,CDSx). - Info: an optional longer description.
- Glyph: the icon that will be associated with the part in the assembly representation. They are taken from the SBOL-visual glyphs. If you want to add more options, reach out via email.
- Left / right overhangs: the 4‑bp sequences that define where this part can connect to its neighbours in the assembly graph.
- Left / right inside: if your parts always include a particular sequence (primer binding, restriction site, etc.) on their left or right end, you can add it here.
- Left / right codon start: if you want to indicate that the overhangs or inside sequences should be in a particular translation frame, you can change the starting codon here.
- Color: the color used to display the part in maps and diagrams. These can be hex colors or named colors, but they must be valid CSS colors. I recommend using this website, which lets you pick several colors at once, and copy their hex codes.
Below is an example illustrating the meaning of the different options for the CDS part.
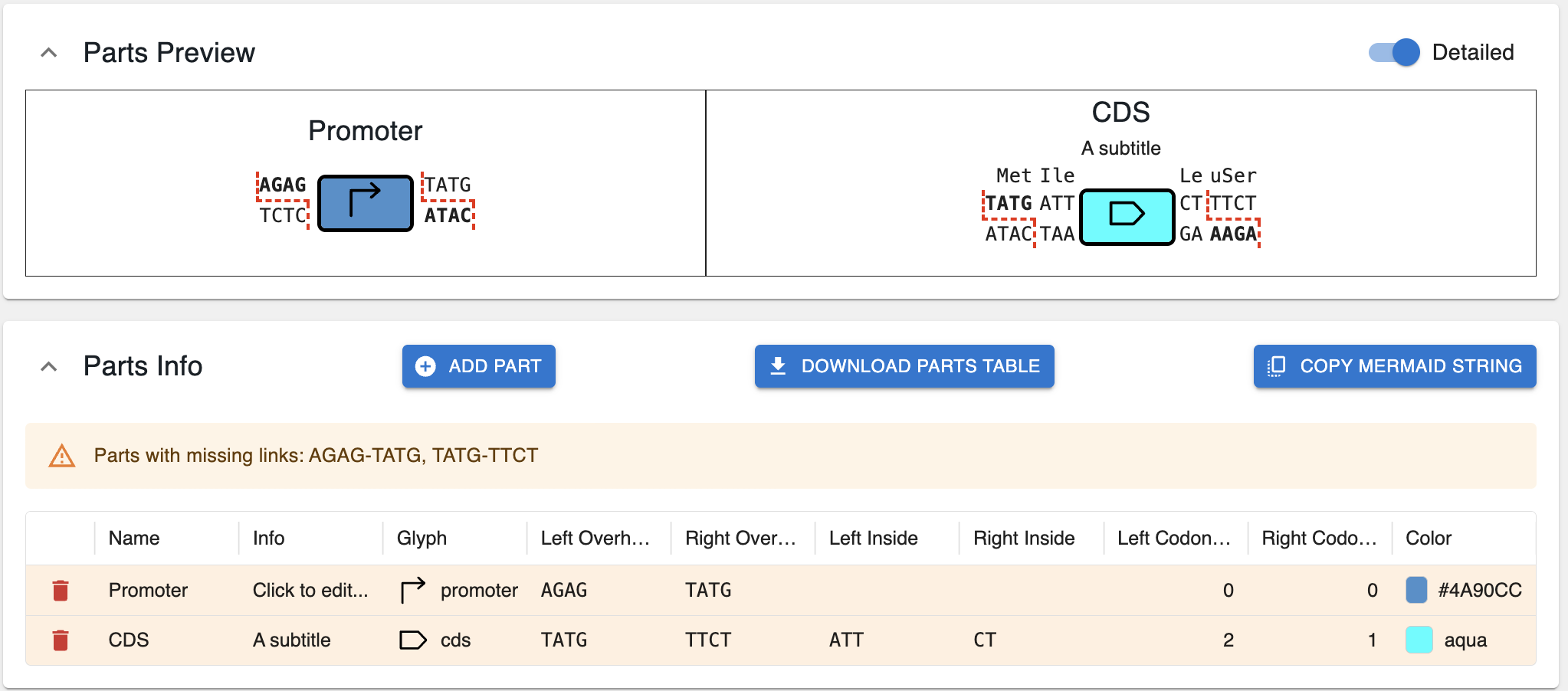
You can also download the parts as a tabular file, if you prefer to edit in a spreadsheet rather than the interface by clicking on Download parts table.
Validation¶
The app checks a handful of things to make sure the syntax is valid:
- All overhangs are connected.
- Only one main circular assemble is defined, which can have internal branching paths, but not two separate cycles.
If you struggle to understand why you are getting an error about cycles, you can click on Copy Mermaid String, and paste it in Mermaid Live Editor to see the graph. That can help identify the problem.
Naming overhangs¶
Some syntaxes do not give a name to the overhangs, such as the MoClo YTK syntax. Others, like GoldenPics give name to overhangs. For instance, the CATG overhang is named 2 and the GCTT is named 3. Therefore, the CDS part that spans CATG-GCTT is also referred to as 2_3.
You can define the naming of overhangs in the Overhang Names section.
General kit information¶
At the top of the page, you can provide general information about the kit. The only necessary part is the name of the kit and the Assembly enzyme (the one that is used to cut the plasmids when assembling them).
You can also provide: * The DOIs of related articles. * Your ORCID (as a submitter of this syntax), and others involved. This is NOT the ORCID of the authors of the related articles, but the ORCID of the person creating the syntax. * The domestication enzyme associated with the kit and domestication overhangs.
Updating linked plasmids¶
For this to work, you must have selected the correct Assembly enzyme (see above section).
Once you have defined your syntax, you may want to check that it makes sense for the plasmids that are associated with it. You can do this by clicking on Update linked plasmids at the bottom of the page.
⚠️ This assumes that you are submitting plasmids, so everything will be treated as a circular sequence (e.g. if you submit fasta, or wrongly annotated genbank files).
Once they are updated, you will see a table with the plasmids, and whether they contain parts that are compatible with the syntax.
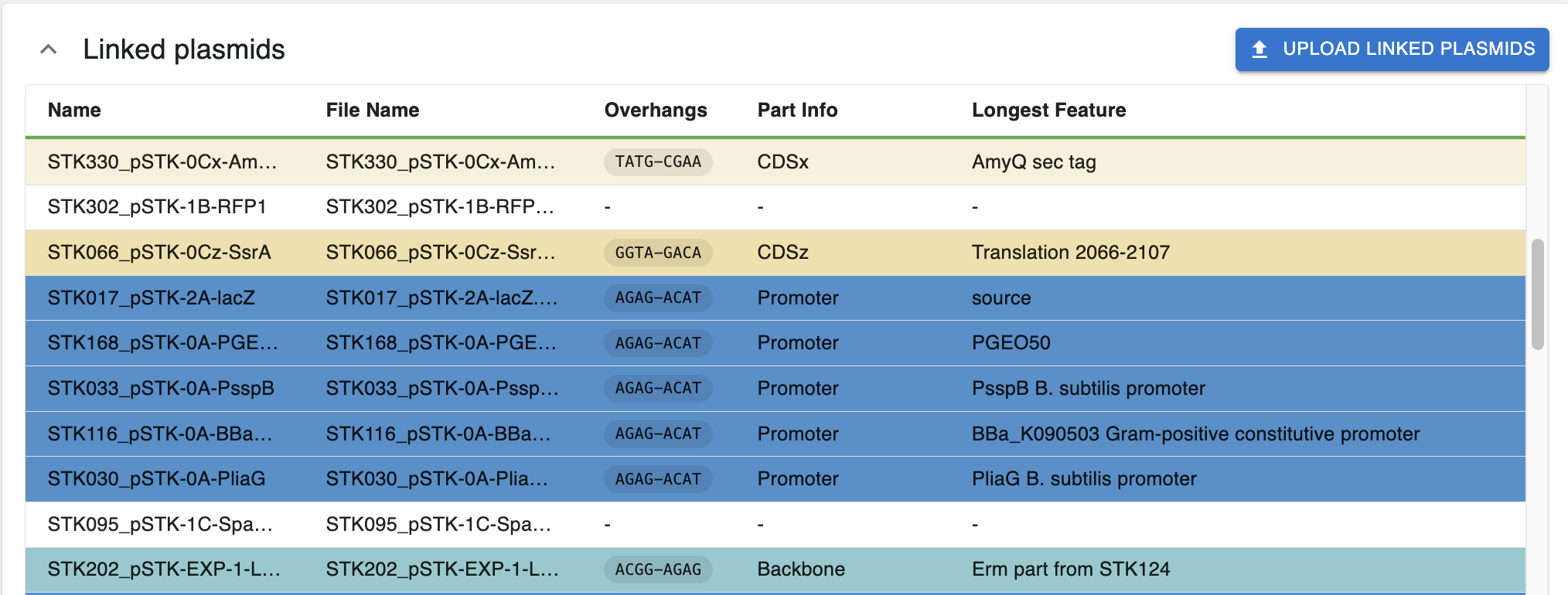
If they are compatible (cut by the assembly enzyme and producing a single part with overhangs compatible with the syntax):
- They are color-coded with the color of the parts in the syntax.
- The column
Part Infodisplays the name of the part based on the syntax. - If the part spans multiple overhangs and it's not named, it will not have a color and say
Spans multiple parts. - If the sequence is annotated, the column
Longest Featuredisplays the name of the longest feature in the restriction fragment that contains the part compatible with the syntax.
Otherwise, they are not coloured. This can be for several reasons:
- Are not cut by the enzyme
- Do not produce overhangs compatible with the syntax when cut.
- When cut by the enzyme, produce more than one part compatible with the syntax.
Using and sharing your syntax¶
- You can download your syntax as a JSON file by clicking the
Download syntaxbutton at the top of the page. - You or others can reload this JSON file to the Syntax Builder to see it or edit it.
- You can update this JSON file to the
Assemblerin OpenCloning to plan combinatorial assemblies with your syntax, see the Assembler documentation for more information. - If you want your syntax to be included in the OpenCloning website, you can create an issue in this GitHub repository or reach out via email.
Feedback and support¶
If you have any feedback or questions, you can reach out via email.