In Vivo Assembly (IVA cloning)¶
What is In Vivo Assembly?¶
In Vivo Assembly (IVA) uses the host cell's homologous recombination machinery to join multiple DNA fragments that share short homologous overlaps at their ends. Instead of assembling the DNA in a tube (like Gibson), fragments are co-transformed and the cell repairs and stitches them together.
- Typical overlaps: 20–60 bp for yeast; bacteria often require longer overlaps and/or recombineering strains.
- Inputs can be: a linearized backbone plus one or more inserts, or multiple fragments that together recreate a circular plasmid.
- Result can be linear or circular, but most workflows aim to recover a circular plasmid after transformation.
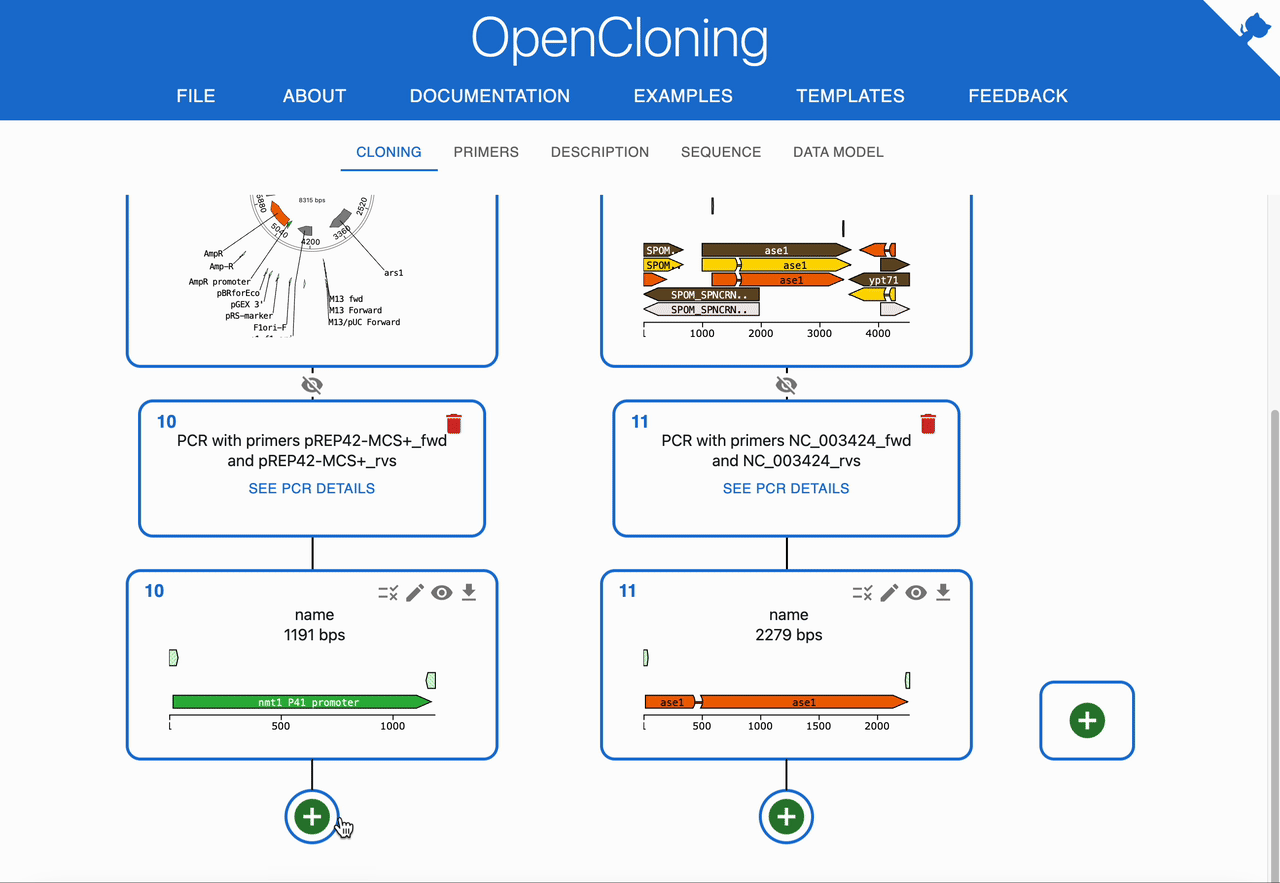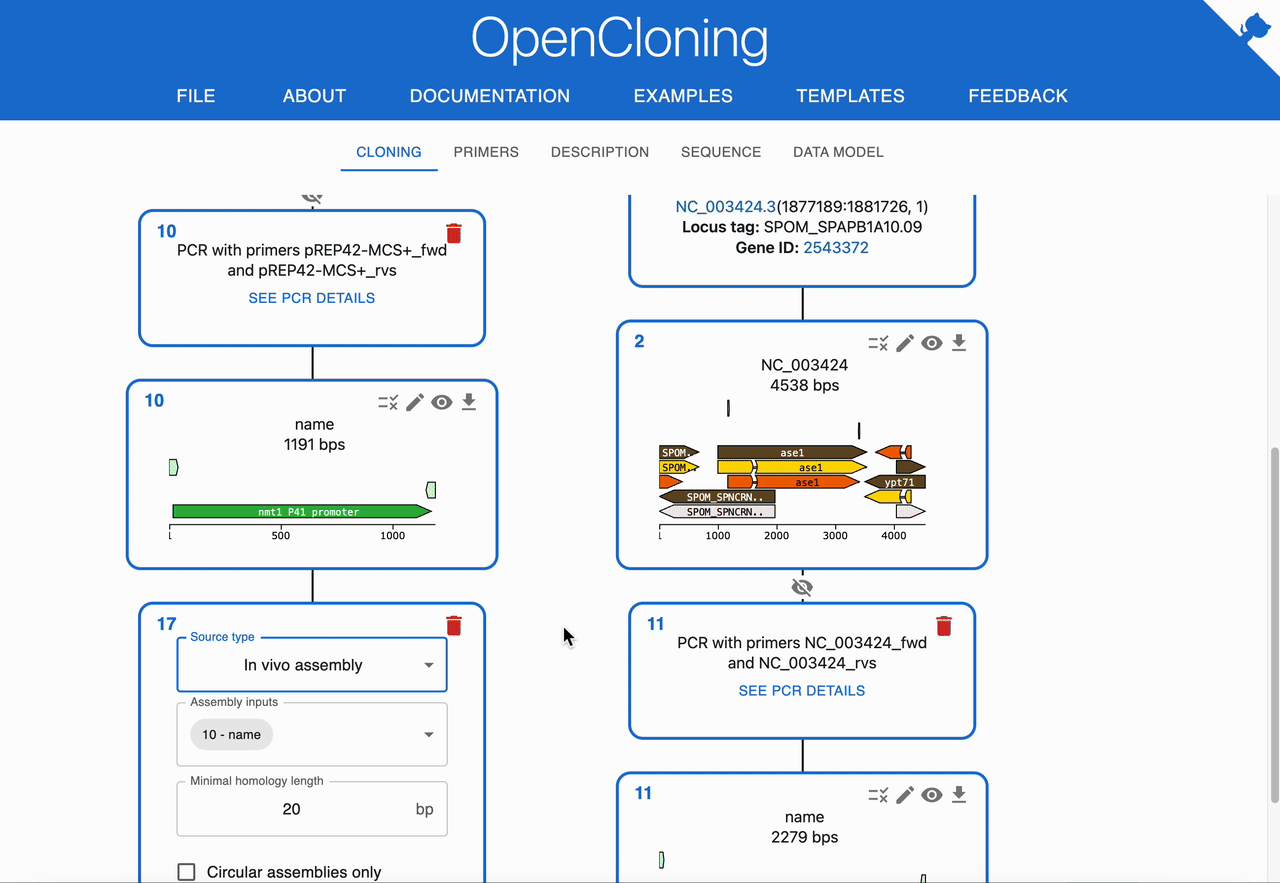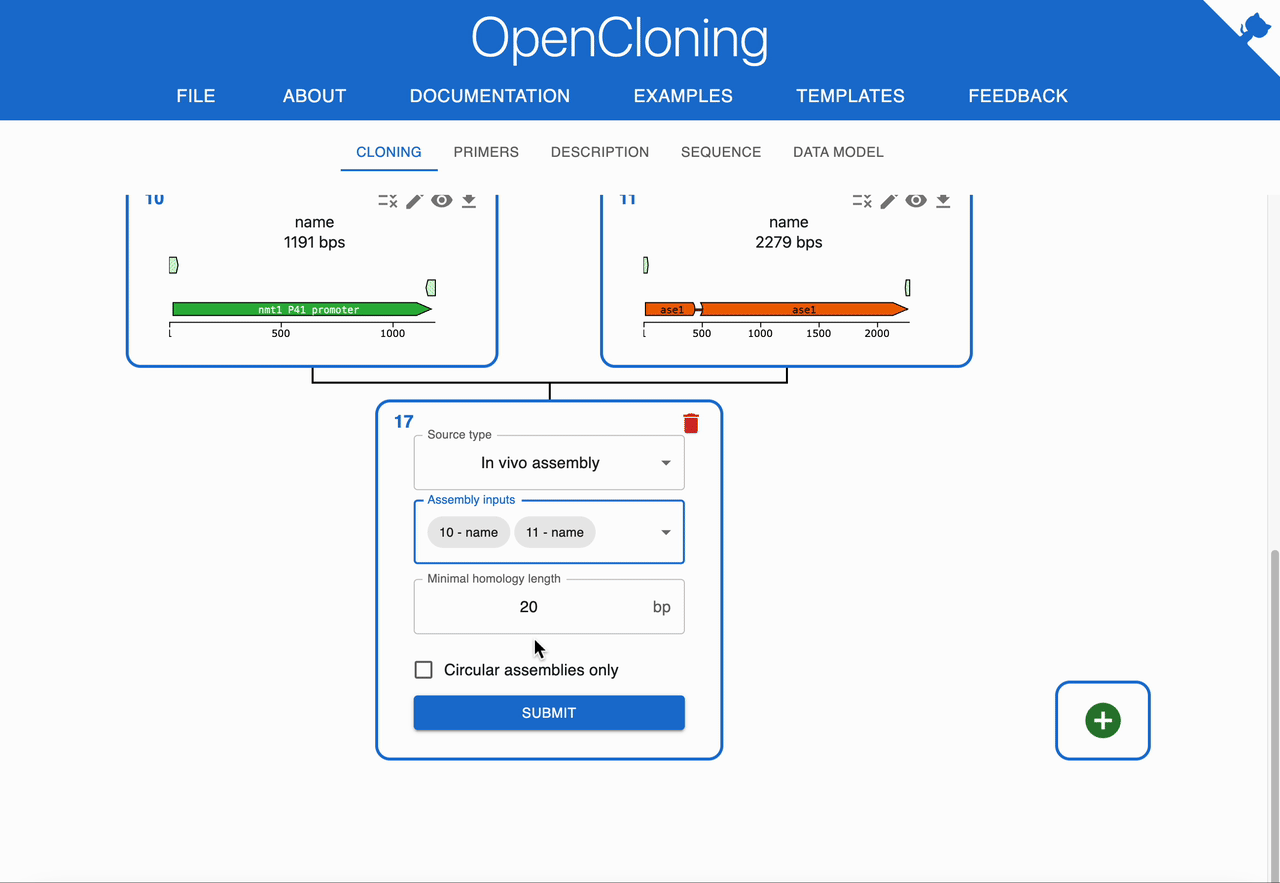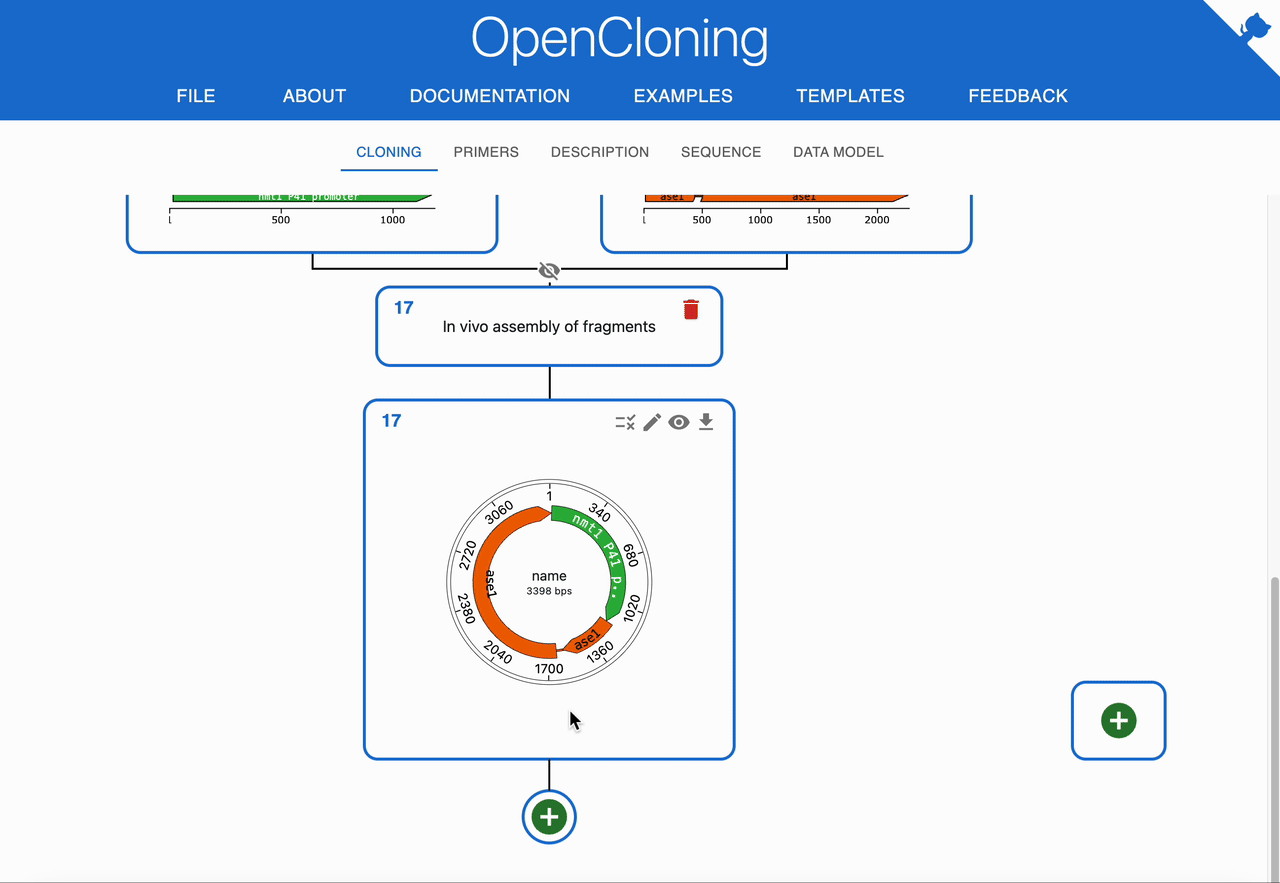
How to plan In Vivo Assembly using OpenCloning¶
Like any other cloning method, click on the plus icon below a sequence in the Cloning tab and select In Vivo Assembly. Then, select the sequences to combine in the Assembly inputs field. OpenCloning will infer valid assemblies by checking adjacent fragments for sufficient homologous overlaps at their ends.

If you need to add overlaps to your fragments by PCR before IVA, see Primer design for Gibson assembly. The process is similar, simply select "In Vivo Assembly" in the primer design form.
Tips and troubleshooting¶
- If you get too many assemblies, increase the minimum required overlap length to reduce spurious joins.
- If you get no assemblies, decrease the minimum overlap length or verify that your chosen fragments actually share terminal homology in the intended orientations.
- For genome integration via homology, use Homologous recombination or CRISPR‑HDR instead.