Gibson Assembly¶
What is Gibson Assembly?¶
- Gibson Assembly is a method to join DNA linear fragments that have shared sequences at their ends.
- It uses an exonuclease that chews back DNA from the 5' end of the fragments, creating sticky ends at both ends of fragments, so that they can be joined together.
- Finally, a DNA polymerase adds the complementary strand of gaps left by the exonuclease, and a ligase removes nicks.

How to plan Gibson Assembly using OpenCloning¶
A Gibson assembly takes as input sequences that already have shared overlaps at their ends. If you need to design primers to add the Gibson shared overlaps via PCR, see Primer design.
To simulate Gibson Assembly, click on the plus icon below a sequence in the Cloning tab and select Gibson Assembly. Then, select the sequences that you want to combine in the Assembly inputs field.
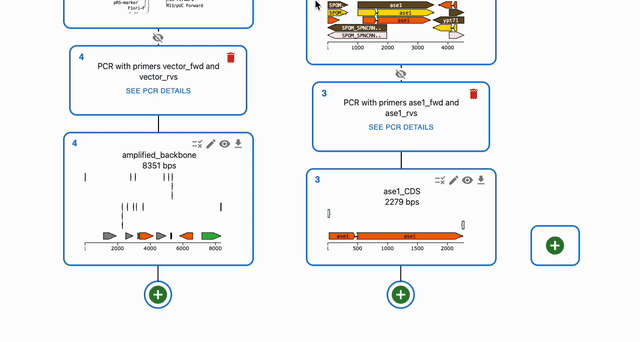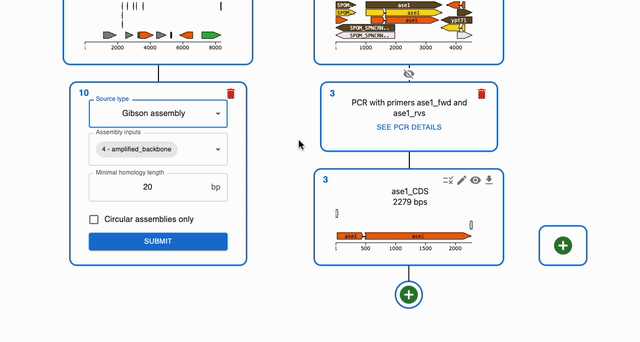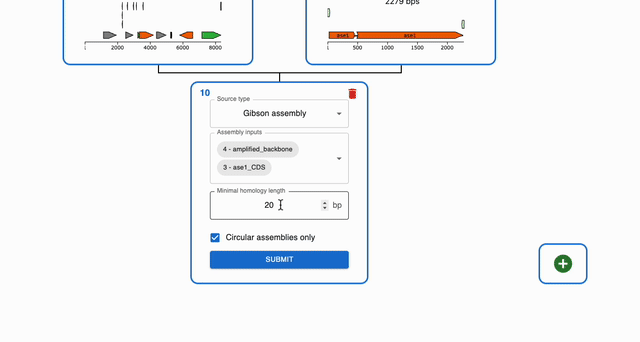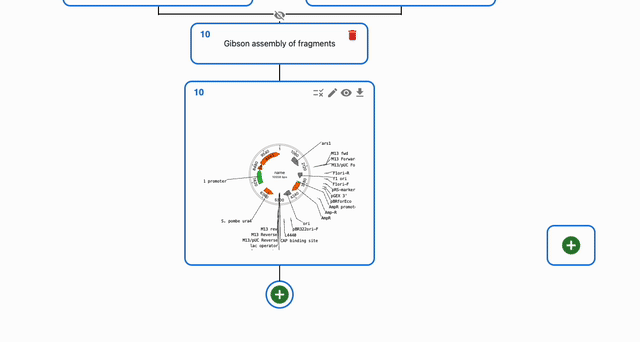
Circularizing a single fragment using Gibson Assembly¶
You can use Gibson Assembly to circularize a single fragment. To do so, don't select any other sequence in the Assembly inputs field. And click on Submit.
You can also design primers for this.